The research of the Pediatric Center of Excellence in Nephrology will seek to generate molecular and spatial maps of every cell type (postal codes) and their gene regulation in healthy and diseased pediatric kidneys. The goal being the creation of a benchmark knowledge base utilized for discovery and validation by students, teachers, researchers, engineers, biologists, clinicians and patients.
We hope our efforts will lead to designing better kidney replacement strategies and provide new insights into therapies aiming to slow decline in kidney function with age or in disease states.
The objective of the pediatric Kidney Single Cell Atlas Project (pKidCAP) is to create spatially resolved single cell molecular maps of pediatric reference and diseased kidneys at several time points across the pediatric lifespan. Discovery of regulatory factors associated with maturity and maladaptive or injury states will not only inform on therapeutic strategies but also in tissue engineering efforts. The cutting-edge technologies being applied include single nucleus RNA-sequencing (snRNA-seq) and chromatin accessibility (snATAC-seq), CUT&RUN, spatial transcriptomics (Visium, CosMx, Xenium) and whole genome sequencing.
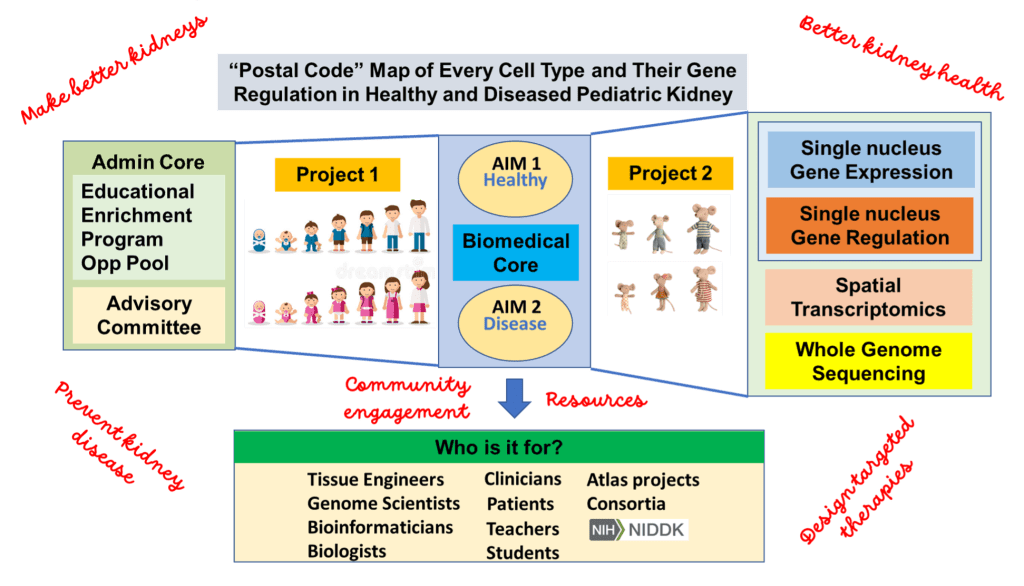
Projects
Project 1
Project 1 of pKidCAP will construct a multimodal atlas of spatially resolved single nucleus gene expression and chromatin state maps with accompanying genomic variants at key stages of pediatric kidney development by using unique and innovatively collected high quality pediatric kidney samples from the Biomedical Core (pKidBIO).
Project 2
Project 2 will investigate the role of a new genetic driver of focal segmental glomerulosclerosis (FSGS). Here single cell datasets, as in Project 1, will be generated at time points approximating the human postnatal development in control mice (for cross-species comparisons) and mouse model of FSGS (to find similarities in gene regulatory circuits with human FSGS).